Synthesis of double stranded oligonucleotides
As well as having the technology to produce a single stranded oligonucleotide Almac also has the capabilities to produce double stranded oligonucleotides.
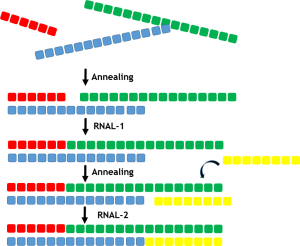
There are a number of different strategies that utilise both single stranded RNAL ligases (ssRNAL) and double stranded RNA ligases (dsRNAL) to accomplish production of the double stranded oligonucleotides. To create a double stranded oligonucleotide a range of short complementary RNA sequences, natural, unnatural or a mixture can be assembled and annealed before using ligase enzymes to join the adjacent fragments as illustrated in the figure below.
The diverse selectAZyme™ RNAL panel facilitates the screening of various strategies to construct double stranded oligonucleotides. A number of approaches are possible, as outlined below, where the optimal enzyme can be identified for each critical step of the process towards the final double stranded product.
Preparation of building blocks
Template independent synthesis of single stranded RNA using Almac selectAZyme ™ single strand RNA Ligase (ssRNAL) and Alkaline Phosphatase (ALKP) panels. Single nucleotide addition can also be performed.


Building block ligation to form double strand RNA
Single stranded pieces can be prepared with ssRNAL then annealed and ligated with dsRNAL. Highly efficient single strand ligations can be determined that reduce the size (and cost) of chemically synthesised building blocks.
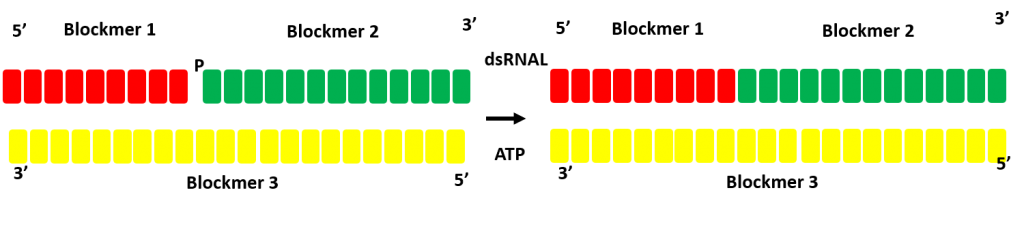

Optimisation of double strand ligation strategy
dsRNAL ligation does not require complete annealing allowing for the optimal enzyme selection for each ligation reaction at scale.
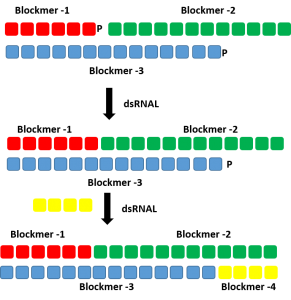

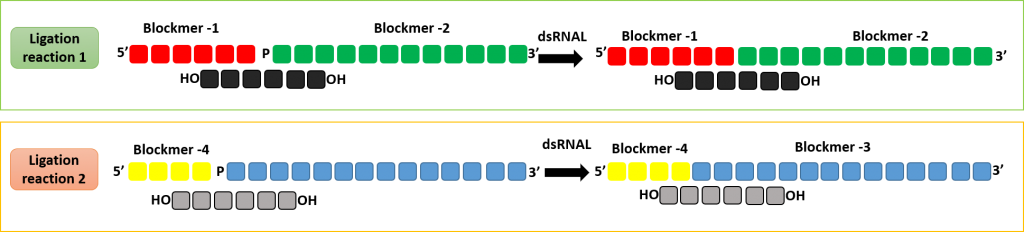
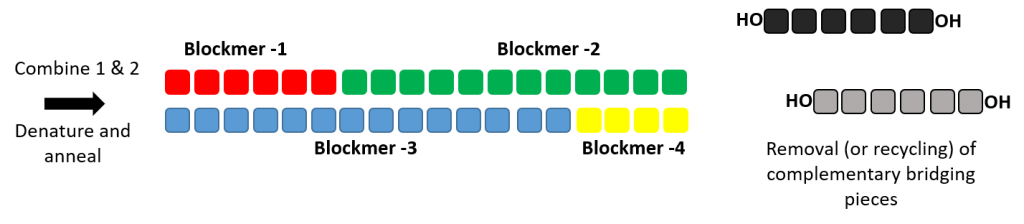
Production of double stranded RNA oligonucleotide using dsRNAL and bridging pieces.
A short complementary bridging piece can be used along with dsRNAL to form a single stranded oligonucleotides. Two ligase reaction can be performed to form the complementary strands of a RNA duplex.