Whole Exome Sequencing (WES) Service
Almac’s high quality, robust WES Next Generation Sequencing (NGS) solution is built on Agilent’s chemistry with sequencing performed on the Illumina NovaSeq or NextSeq technology.
Almac’s wet lab optimised workflow generates comprehensive WES data from multiple sample types, including more challenging Formalin Fixed Paraffin Embedded (FFPE) material. The platform covers 99.7% of coding exons with readily interpretable raw sequencing data for use in biomarker discovery and retrospective clinical investigation.
The dual DNA and RNA extraction protocol facilitates simultaneous generation of WES and RNA Exome data from a single FFPE sample.
For Research Use Only.
Almac Diagnostic Services has received R&D funding from Invest Northern Ireland and the European Regional Development Fund to support the development of this service. For more information click here.
Robust Almac WES Data Generation Protocol:
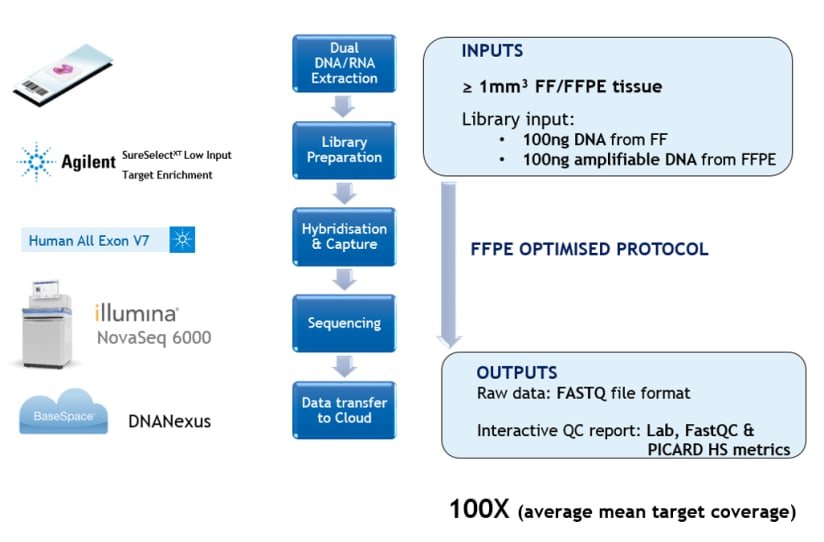
Technical Specification and Performance Validation:
Almac analytical validation data confirms the highly accurate, repeatable and reproducible performance of the WES service.
For full technical specification & further validation information download the fact sheet.
Benefits:
- QC guarantee of 100X mean target coverage across sample cohort ensured comprehensive downstream analysis and data interpretation.
- Protocol optimised for challenging samples including FFPE material.
- Dual DNA & RNA extraction protocol enables generation of WES & and RNAseq data from a single sample.
- A comprehensive variant detection platform, covering 213,994 coding exons.
- Machine-learning based bait design of SureSelect Human All Exon V7 ensures superb streamlined coverage of variants across exons.
- Interactive QC report enables rapid and comprehensive sample quality interrogation across cohort.
- Raw sequence data delivered in FASTQ format and compatible with the majority of bioinformatics pipelines.
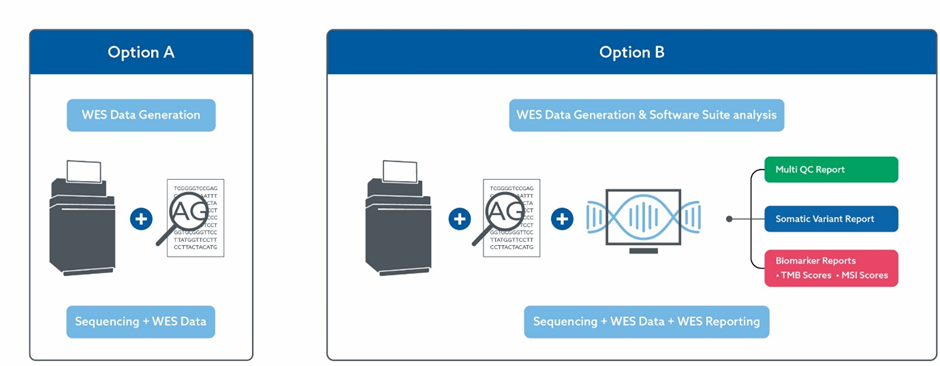
Options
Clients can choose the following options:
- OPTION A Almac WES data generation service only
- OPTION B Almac WES data generation service coupled with the Almac WES Software Suite analysis

Almac WES Software Suite
Almac Diagnostic Services’ Data Science Team have further developed a bespoke proprietary Almac WES Software Suite analysis tool to complement the Almac WES Data Generation Service. The WES Software Suite has been innovatively designed to “clean-up” variant calls to allow for the facilitation of high-quality downstream analysis.
The software suite consists of a tumour-normal and tumour-only somatic variant detection workflow. The key tumour-only capability enables accurate determination of TMB from FFPE tumour samples without a matched normal.
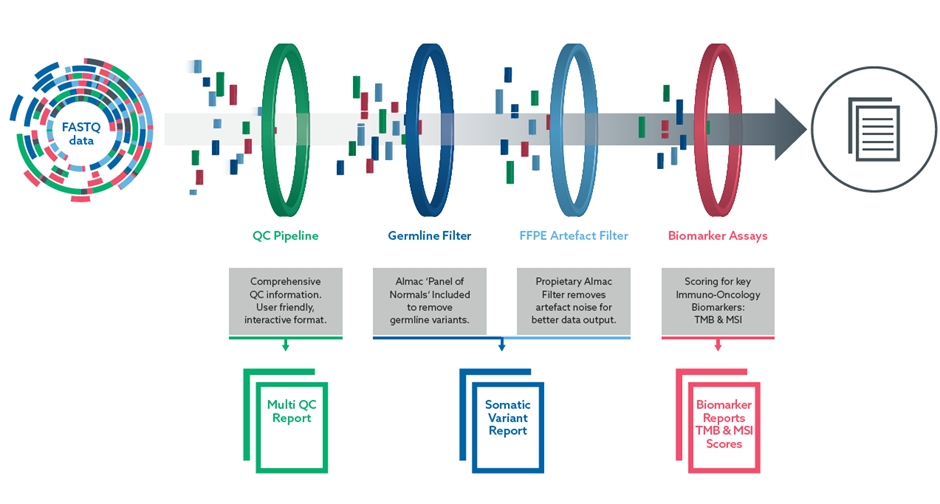

| Filters & Reports | Features & Benefits |
| Multi QC Report | A comprehensive QC report, enabling clients to review data in a html interactive format compared to a standard txt file. A ‘user friendly’ design for more rapid interpretation. |
| Tumour-normal (TN) pipeline | Gold-standard somatic small variant detection with minimal noise. |
| Tumour-only (TO) pipeline | Accurate somatic small variant detection without cost of additional sample processing & analysis costs. |
| Germline filter (Including Panel of Normals) | Removes background “germline” variants to identify ONLY somatic variants without need for processing a normal sample. |
| FFPE artefact filter | Removes technical artefacts inherent in formalin-fixation process to enrich for clinically relevant variants. |
| TMB and MSI scoring | Critical biomarker information for I-O studies. |
Abstract
View Almac’s abstract and poster at AACR on “A machine learning approach to detect FFPE artefacts leads to accurate estimation of tumor mutational burden from sequencing data without a matched normal.”
Contact us:
Interested in Almac’s Whole Exome Sequencing (WES) Service? Submit your details below:



