Whole Genome Sequencing (WGS) Service
A high quality, robust Next Generation Sequencing (NGS) solution built on Roche’s platform. Almac’s wet lab workflow generates comprehensive WGS data from blood.
The platform covers coding and non-coding regions with readily interpretable raw sequencing data for use in biomarker discovery and retrospective clinical investigation.
QC guarantee of 30X mean target coverage across sample cohort ensures comprehensive downstream analysis and data interpretation.
For Research Use Only
Robust Almac WGS Data Generation Protocol
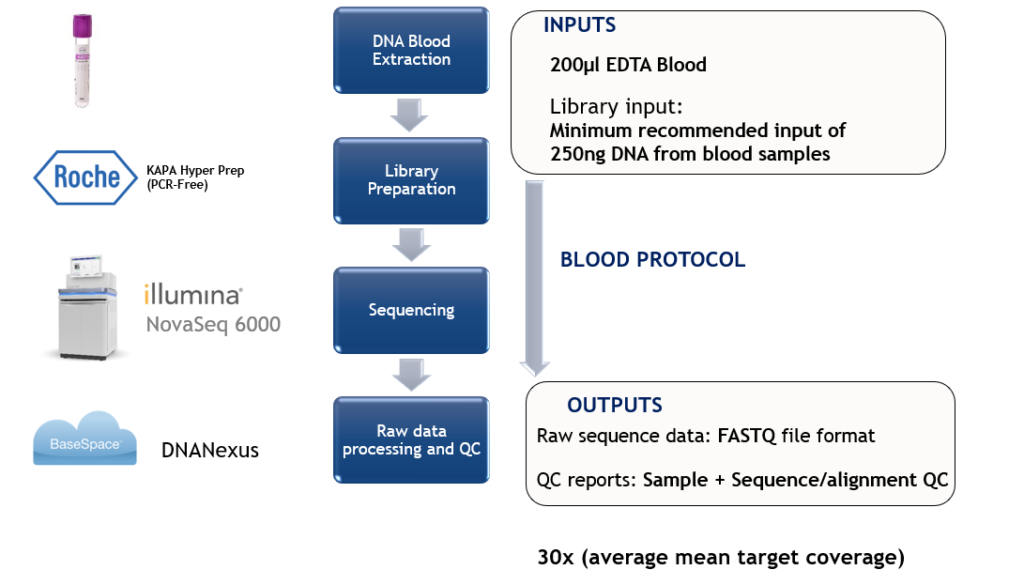
Technical Specification and Performance Validation:
Almac analytical validation data confirms the highly accurate, repeatable and reproducible performance of the WGS service.
For full technical specification & further validation information download the fact sheet.
Benefits:
- QC guarantee of 30X mean target coverage across sample cohort ensures comprehensive downstream analysis and data interpretation
- Protocol optimised for blood samples
- Interactive QC report enables rapid and comprehensive sample quality interrogation across the cohort
- Raw Sequence data delivered in FASTQ format, compatible with the majority of bioinformatics pipelines
- Germline variant discovery service: filtered and annotated germline variants delivered as standard VCF files compatible with majority of downstream interpretation tools
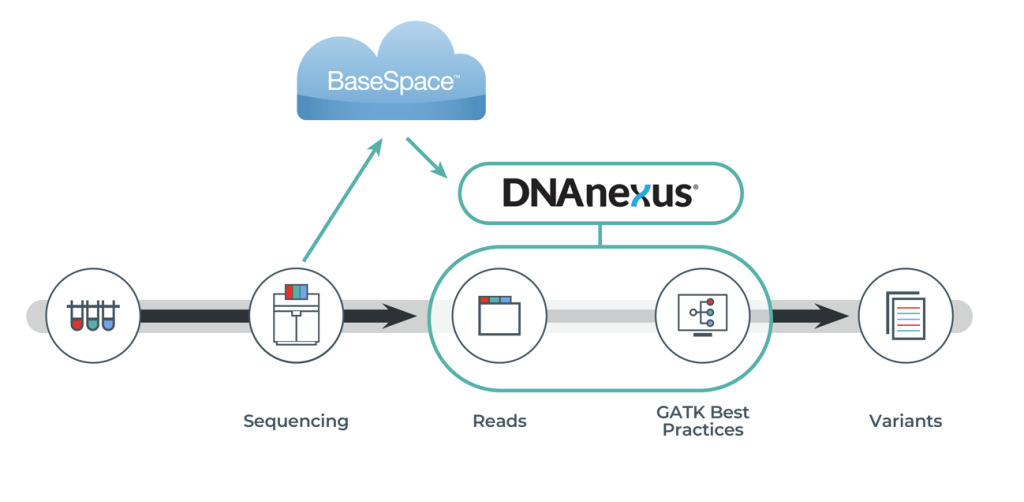

Almac provides a germline variant discovery service, including standard variant QC filtering and annotation steps.
Outputs:
- Raw sequence in FASTQ format
- Interactive HTML sequence/alignment QC report
- Germline variant calls delivered as VCF files compatible with majority of downstream interpretation tools- Raw and Filtered + annotated
- Custom downstream variant analysis options are also offered.



