What’s new?
-Additional Content – 16 NEW Gene Expression Signatures across the Hallmarks of Cancer.
-Improved Functionality – New Web Access to view and interpret your data.
-Simplified Reporting – One Interactive Dashboard View. No need for multiple files and folders.
What is claraT ?
claraT is a unique software-driven solution, classifying biologically relevant gene expression signatures into a comprehensive, easy-to-interpret report.
The claraT * Report
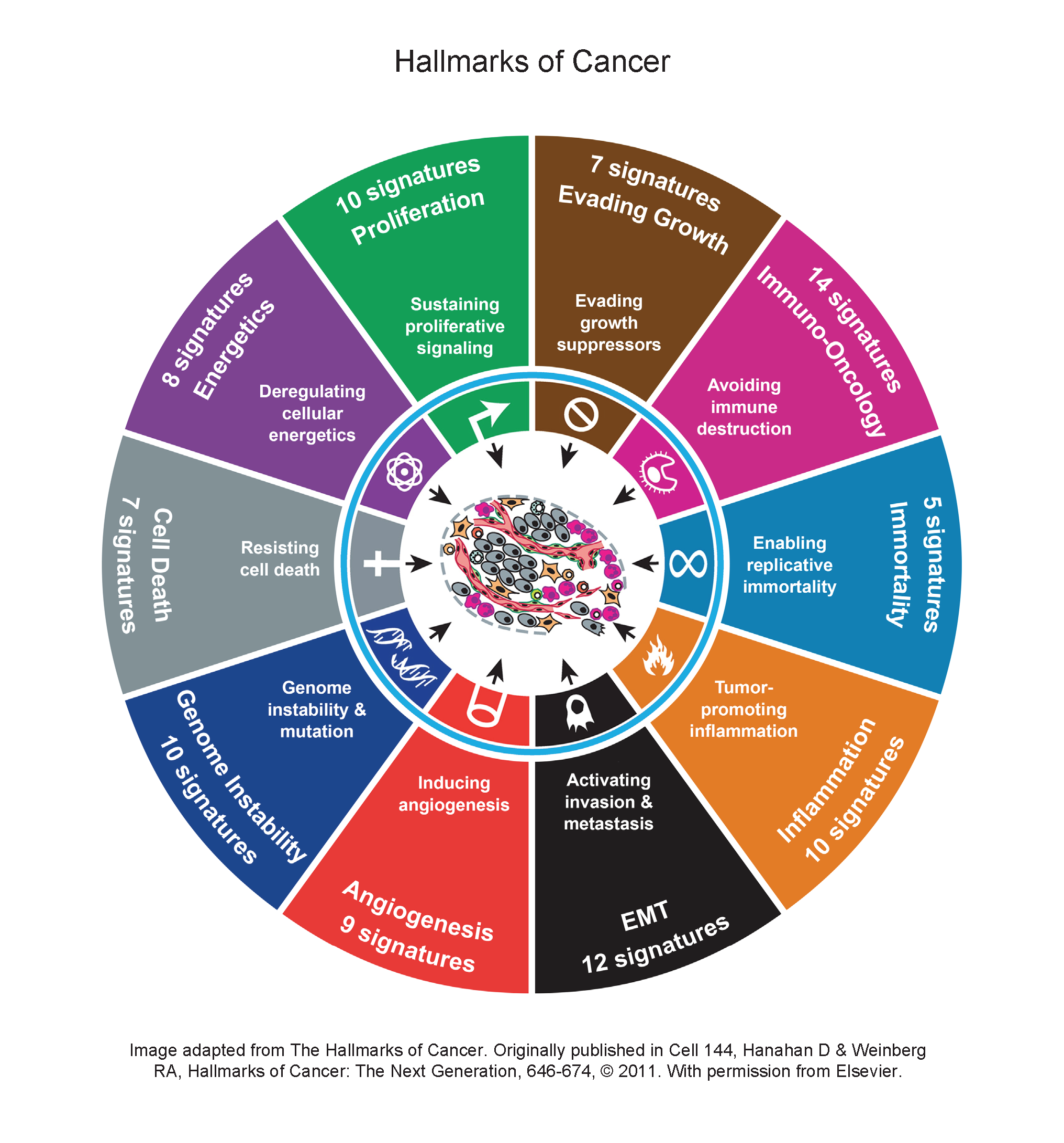
claraT classifies multiple publicly available gene expression signatures and single gene targets linked to multiple key biologies, alongside Almac’s own proprietary assays, according to the 10 Hallmarks of Cancer.
Clients are provided with a unique, interactive report that allows the easy visualisation of the key discriminating biologies within either a large cohort or an individual tumour sample.
Supplementary files are also included in the report and are hyperlinked within each profile for additional data functionality.
A pan-cancer solution, based on a powerful bioinformatics pipeline, automatically generating the claraT report from raw gene expression data utilising Almac RNA-Seq Solutions.
* Note: claraT is for research use only (RUO) and is not to be used for diagnostic or prognostic purposes, including predicting responsiveness to a particular therapy.
Contact Us
claraT benefits:
Maximises the understanding of your dataset
Provides readouts for the most relevant published gene expression signatures
Standardises comparisons between different datasets
Highlights consensus between multiple gene expression signatures
Provides extensive supplementary data for innovative analysis
claraT Report - Facts:

First conceptualised by Douglas Hanahan and Robert Weinberg in 2000 and subsequently updated in 2011, the Hallmarks of Cancer have become a paradigm within modern cancer research.
The hallmarks help explain the complexity of cancer cells and describe the processes that occur allowing cancer cells to proliferate and grow if unchecked.
The claraT report utilises the Hallmarks of Cancer concept of targetable biologies to help facilitate biomarker discovery. claraT embodies the tenets set out by the authors, intelligently simplifying the complex biologies of cancer within key hallmarks.