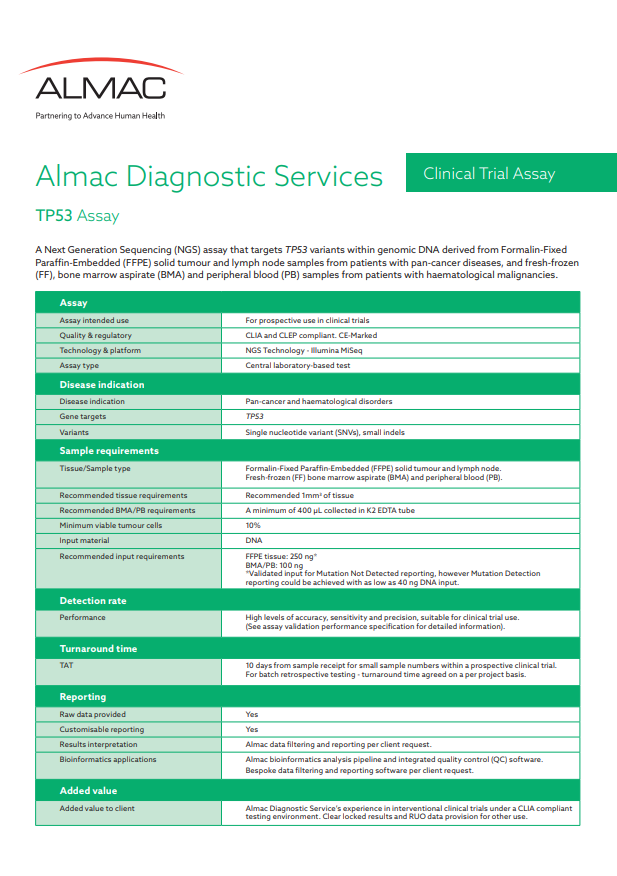
TP53 Clinical Trial Assay
Almac has developed a Clinical Laboratory Improvement Amendment (CLIA) compliant analytically validated Next Generation sequencing (NGS) assay for the TP53 tumour suppressor gene, which is linked to many cancers. This clinical trial assay (CTA) is intended to detect germline and somatic DNA aberrations enabling determination of molecular eligibility to facilitate clinical trial enrolment. The assay utilises a dual amplicon workflow which can effectively determine true variants from FFPE artefacts, and unique molecular identifiers (UMIs) which can identify sequencing errors. These features taken together provide a highly accurate assay for both mutation detection and wildtype calling.
Assay background
The Almac TP53 CTA is a qualitative NGS assay intended to detect genomic aberrations. It is validated for use with Formalin-Fixed Paraffin-Embedded (FFPE) solid tumour and lymph node samples, from patients with pan-cancer diseases, as well as fresh-frozen (FF), bone marrow aspirate (BMA) and peripheral blood (PB) samples from patients with haematological malignancies. The assay detects single nucleotide variants (SNVs) and small indels from DNA, enabling samples to be accurately classified as Mutation Detected or Mutation Not Detected.
Analytical validation
The assay has been analytically validated in accordance with CLIA and New York State Department of Health Clinical Laboratory Evaluation Program (CLEP) requirements and is suitable for interventional use within clinical trials, such as for determining molecular eligibility. The assay is designed and developed in accordance with ISO 13485:2016 (Medical Devices – Quality Management Systems – Requirements for Regulatory Purposes) and ISO 14971:2019 (Medical Devices – Application of risk management to medical devices) and comprises a validated bioinformatics analysis pipeline and integrated quality control (QC) software which conform to IEC 62304:2006+A1:2015 (Medical Device Software – Software life-cycle processes) and ISO 14971 standards.
Bespoke data reporting option
Almac’s Data Science team experts can also develop bespoke data filtering, annotation and reporting software on a per-project basis for clients.
Key benefits
- Analytically validated CTA for prospective testing of FFPE tissue and FF BMA/PB samples
- Full coding coverage of all TP53 isoforms
- Detection of single nucleotide variants and small indels
- Watson & Crick DNA strand coverage and incorporation of unique molecular identifiers (UMIs) and stringent coverage QC resulting in high-confidence Mutation Detected & Mutation Not Detected status
- Rapid turnaround time
- Access to raw data
- Comprehensive reporting and interpretation services
ctDNA potential
- UMI based technology can enable further development of the assay for ctDNA samples.
Contact us below to initiate a discussion.
Key advantages of Almac TP53 single gene assay vs panel approach
- Equivalent Input
- Lower Cost
- Higher Sensitivity
- Greater Coverage
- Faster Turnaround Time (TAT)
Comparison of Almac TP53 single gene assay vs leading panel providers:
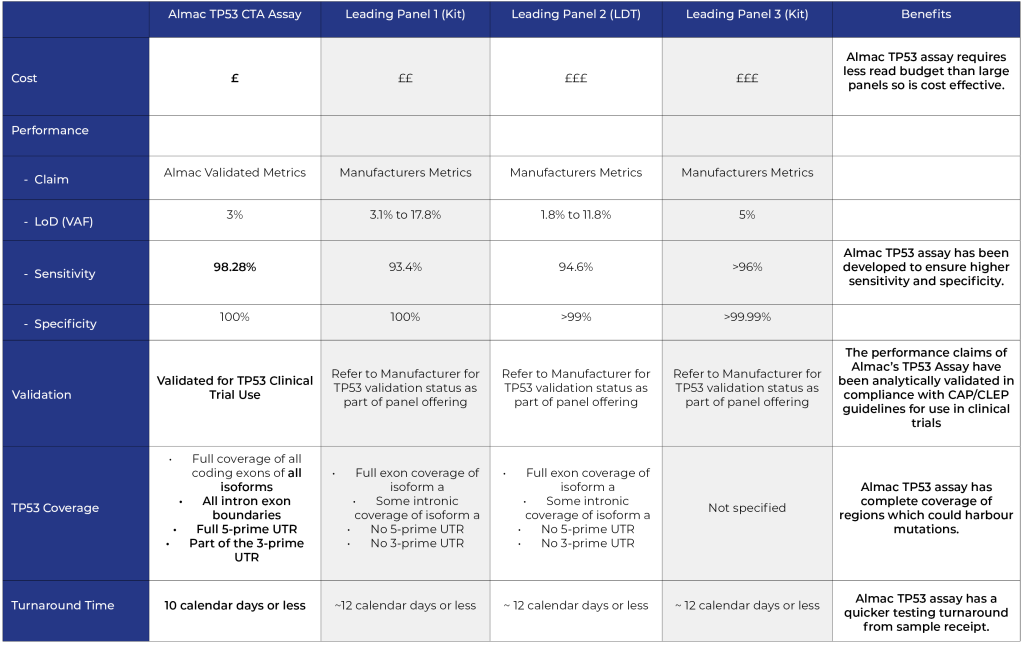
NOTE: Comparison metrics correct as at February 2023
*By submitting your information you acknowledge that you have read the privacy statement and you consent to our processing the data in accordance with that privacy statement. We may, from time to time, send you material relevant to your interests. If you change your mind at any time about wishing to receive material from us, you can send an email to [email protected]. Every email we send you will also include an unsubscribe link so you can unsubscribe from our marketing list.