The increasing uptake of RNA-Seq as the technology of choice in biomarker discovery

Dr Nuala McCabe
Biomarker Research Manager, Almac Diagnostic Services

RNA biomarkers – a largely untapped, but beneficial resource
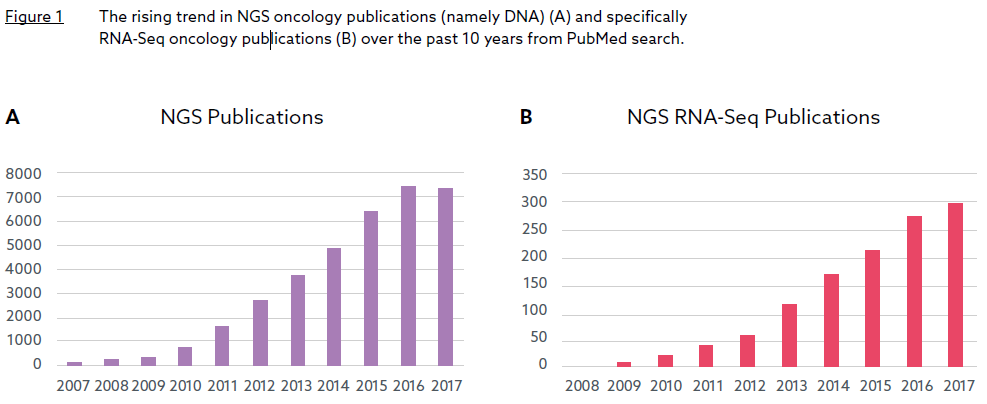
Next generation sequencing (NGS) has evolved as a valuable tool for biomarker discovery and development. This new era of research into biomarker discovery has largely focused on DNA based biomarkers rather than RNA based biomarkers and is exemplified by the number of publications in the NGS field, mostly DNA related (Figure 1A). To date, RNA has been a relatively untapped resource for biomarker discovery. However, within the past 8-10 years, there has been a significant rise in the number of NGS publications featuring RNA-Sequencing (RNASeq) (Figure 1B). This change has been driven by the realisation that researchers cannot solely rely on DNA aberrations to understand tumour biology but also need other omics data such as RNA profiling to capture a more comprehensive view.
This suggestion has recently been highlighted in a New York Times article entitled “The Search for Cancer Treatment Beyond Mutant-Hunting” by Siddhartha Mukherjee (author of the book “The Emperor of All Maladies: A Biography of Cancer”) who suggested “mutations within a cancer cell certainly carry information about its physiology – its propensity for growth, its vulnerabilities, its potential to cause lethal disease – but there’s a world of information beyond mutations.” He continues “What if the ‘really clinically useful information’ lies within these domains – in the networks of normal genes co-opted by cancer cells, in the mechanisms by which they engage with their host’s immune system or in the metabolic inputs that a cell needs to integrate in order to grow? At the annual meeting of the American Society of Clinical Oncology (ASCO) this year [2018], it was this altered – and more expansive – vision of precision cancer medicine that was on display. ” It should be noted that several commercial RNAbased biomarkers have already been developed for prediction of outcomes in cancer including Agendia’s – Mammaprint®, Genome Dx’s – Decipher®, and Genomic Health’s – OncoType DX® assays, although these are microarray and q-PCR based rather than using NGS.
The evolution of RNA-Seq
Traditionally cDNA microarray technology has been the primary method of choice for gene expression profiling. However, with the continued advancement of NGS technology, RNA-Seq is rapidly emerging as the principal discovery tool. Unlike microarrays which are limited in their design to the detection of known transcripts, RNA-Seq has the potential to detect novel transcripts and structural variants such as alternative splicing events and gene fusions and can also identify allele-specific expression and disease-associated single nucleotide polymorphisms (SNP). Furthermore, RNA-Seq technology also avoids the hybridization based issues associated with microarrays and demonstrates a broader dynamic range (>8,000 fold), with low background signals. For differential gene expression analysis, RNA-Seq demonstrates superior levels of accuracy in the quantification of highly expressed genes and sensitivity in lowly expressed genes when compared to microarrays. Multiple platforms are currently available for RNASeq, including Illumina HiSeq and NoveSeq platforms and ThermoFisher Ion Torrent Personal Genome Machine.
The challenges of RNA-Seq
Formalin-fixed paraffin-embedded (FFPE) samples are a valuable resource for biomarker studies since they are routinely collected in clinical practice. However formalin fixation presents certain challenges both from a technical and bioinformatics perspective. This is mainly due to chemical modification, crosslinking, and general degradation of the RNA, particularly in samples over 6 months old. However, with the advancement of sequencing chemistries and processes for RNA-Seq analysis, optimized workflows (e.g. Illumina TruSeq® RNA Exome) are now available with robust quality control steps both in the laboratory and throughout data analysis, incorporating advanced algorithms which can account for FFPE specific effects. The scale and complexity of the data generated with RNA-Seq also harbours significant challenges for biomarker discovery. Interpretation of RNA-Seq data requires sophisticated and powerful computational programs with novel tools emerging constantly that require performance validation. A wealth of mature tools exist to meet basic requirements (e.g. applications hosted on Illumina BaseSpace Sequence Hub). Some challenges, however, remain where there is scope for more advanced algorithms such as differential gene expression analysis, detection of fusion genes, alternative splicing, and variants detection.
The opportunities of RNA-Seq
Despite the various challenges of using RNA-Seq for biomarker discovery and development, there are numerous opportunities. Gene expression based biomarkers capture several molecular pathways and are more dynamic than DNA based biomarkers. They may therefore better reflect the underlying biology of disease, over time and with treatment
“The obsession with DNA mutations alone is short sighted. Utilisation of RNA signatures is key to recognizing stratified medicine.”- Professor Tim Maughan CRUK/MRC
RNA-Seq allows for the discovery and validation of a large number of gene expression signatures simultaneously from a single tumour sample (from as low as 20-50 ng RNA input), which may enable better treatment stratification through reflecting multiple biologies. In addition, the ability to understand parallel biological pathways guides potential combination drug therapies and aligns with the recent move in clinical trials towards basket and umbrella approaches. The release of public databases of expression data from collaborative programmes such as The Cancer Genome Atlas (TCGA) has facilitated the identification and in silico validation of many potential gene expression Biomarkers for clinical trial use.

The RNA technology market landscape
In my opinion, four main categories of commercial RNA expression profiling exist. The first of these are the service providers for larger whole transcriptome panels Almac, Q2 Solutions and PGDx for example, providing comprehensive sequencing analysis. However, this is a standard service offering, where clients may require additional bioinformatics expertise for data analysis and interpretation. In addition to these whole transcriptome offerings, there are many curated gene panels (some with and some without a specific report) available for biomarker development including the HTG Molecular Panel and Nanostring Panels. These panels have the advantage of a shorter turnaround time, are more focussed on biologies and produce less data for analysis and interpretation. For example, Cofactor Genomics provide an immuno-oncology (IO) only offering with a patient report for instant interpretation. However, these targeted approaches have limitations as they do not provide a comprehensive picture of the global tumour biology. The final category is whole transcriptome analysis with a customised biologically interpreted report such as the Almac claraT proprietary solution (see below). This approach allows the simultaneous application of multiple biomarker assays that represent the hallmarks of cancer, providing the rapid generation of standardised data and saves considerable bioinformatics resource.
claraT– Almac’s unique gene expression report
To solve the complex RNA-Seq data challenge, Almac has developed claraT*- A unique software-driven solution, classifying biologically relevant gene expression signatures into a comprehensive, easy-to interpret report. claraT helps simplify RNA-Seq data for biomarker discovery and translational research. A pancancer solution, based on a powerful proprietary bioinformatics pipeline, automatically generating the claraT report from raw gene expression data utilizing the Almac optimized RNA Exome Panel. The claraT report covers key biologies within the Hallmarks of Cancer, and to date includes 35 gene expression signatures, 30 drug targets and 1,641 single genes relevant to cancer biology.
claraT – Whitepaper:
“Evaluation of the claraT Total mRNA Report In an RNA-Sequencing dataset from malignant melanoma cancer patients.”
In this whitepaper the application of claraT to an RNA-Sequencing dataset of malignant melanoma cancer patients treated with immune checkpoint therapy is described.
In Conclusion
To conclude, RNA Sequencing (RNA-Seq) is an increasingly popular technology for biomarker discovery in cancer research. High-throughput RNA-Seq produces large quantities of data that can reflect multiple biologies but requires complex computational bioinformatics pipelines to enable interpretation. Novel software approaches such as the Almac claraT solution can bring a standardised, rapid approach to analysis of this data for the purposes of biomarker discovery and application.
*claraT is for research use only (RUO) and is not to be used for diagnostic or prognostic purposes, including predicting responsiveness to a particular therapy.
The information contained in this article is accurate, to the best of the author’s knowledge, as of 1 February 2019.
claraT TM is a trade mark of Almac. Other trade marks referenced are those of third parties.
Download file